|
Ali Cakmak, Ph.D. |
| Home |
Bio |
Research |
Publications |
Projects |
Teaching |
Photography |
Tools and Algorithms for Multi-omics Supported Personalized Treatment Recommendation, Drug Repositioning, and Drug Target Discovery
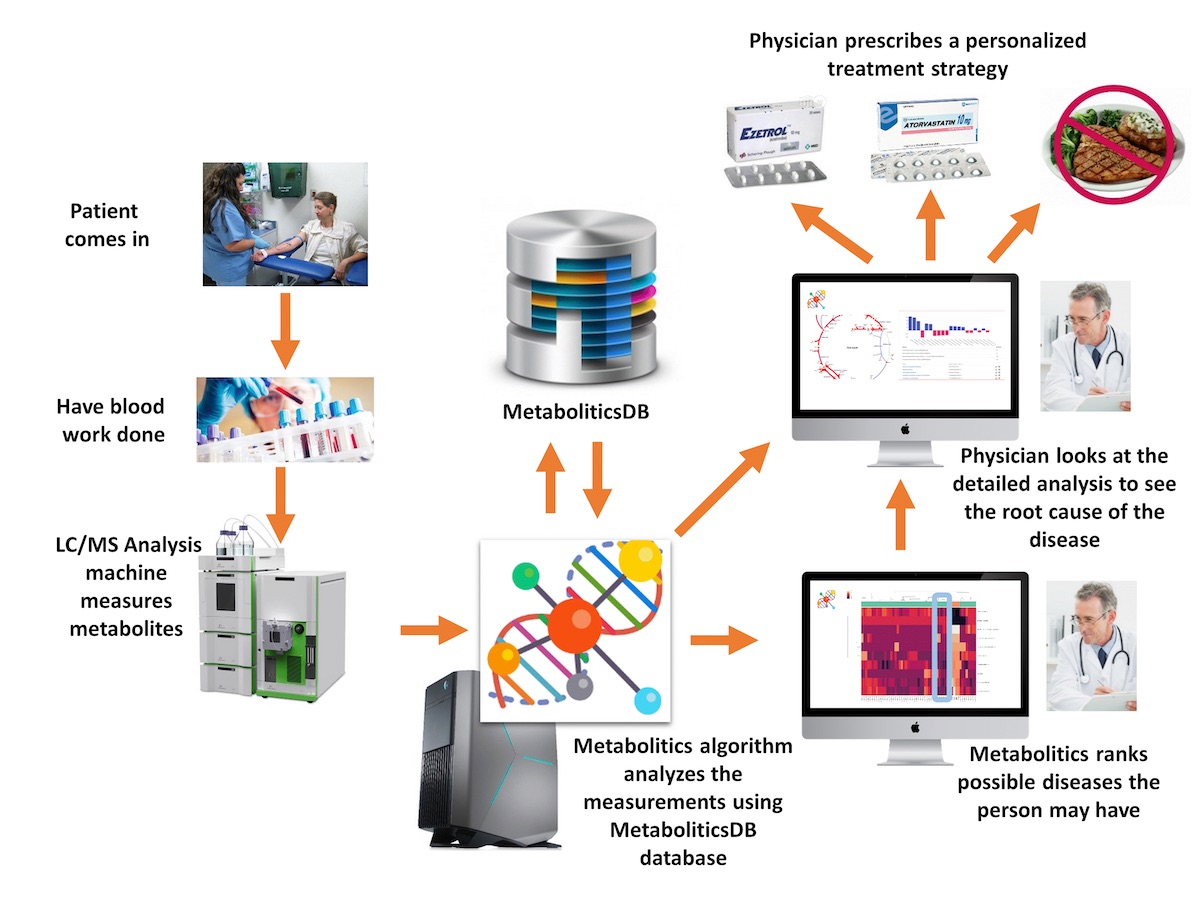
Increasing availability of high throughput omics data at decreasing cost has led to a rapid progress in understanding the mechanisms of diseases. Such developments offer many opportunities for personalized and precision medicine practices, as well as drug development. This project focuses on the development of tools and methods for the integrated and individual analysis of different omics data in terms of changes in human metabolism, and later the exploitation of such analysis results for personalized treatment selection and the development of new drug options. The proposed project will be based on the metabolomics data analysis method and related tools that have been developed as part of a previously completed TÜBİTAK project. In summary, the aim of this project, in the first level, is to develop methods to perform combined and separate analysis of different types of omics data that are obtained from the same individual in terms of metabolic perturbations. Next, in the second level, the project’s goal is to utilize these analysis methods and associated results to develop (a) a personalized drug recommendation tool based on omics data of an individual, (b) a tool to propose candidate drugs for repositioning, and (c) another tool discover new drug targets. Our aim in the third layer is to make these methods available to researchers and clinicians as web-based and database-supported interactive tools. In addition, a select subset of drugs that are predicted for repositioning will be studied in wet lab on appropriate cell lines for in-vitro verification.
Funding Institution: The Health Institutes of Turkey (TUSEB TA-01) (Role: PI, Duration: 36 Months, 2020 - 2023)
 Algorithms and Tools for Computational Modeling of Metabolomics Data
Algorithms and Tools for Computational Modeling of Metabolomics Data
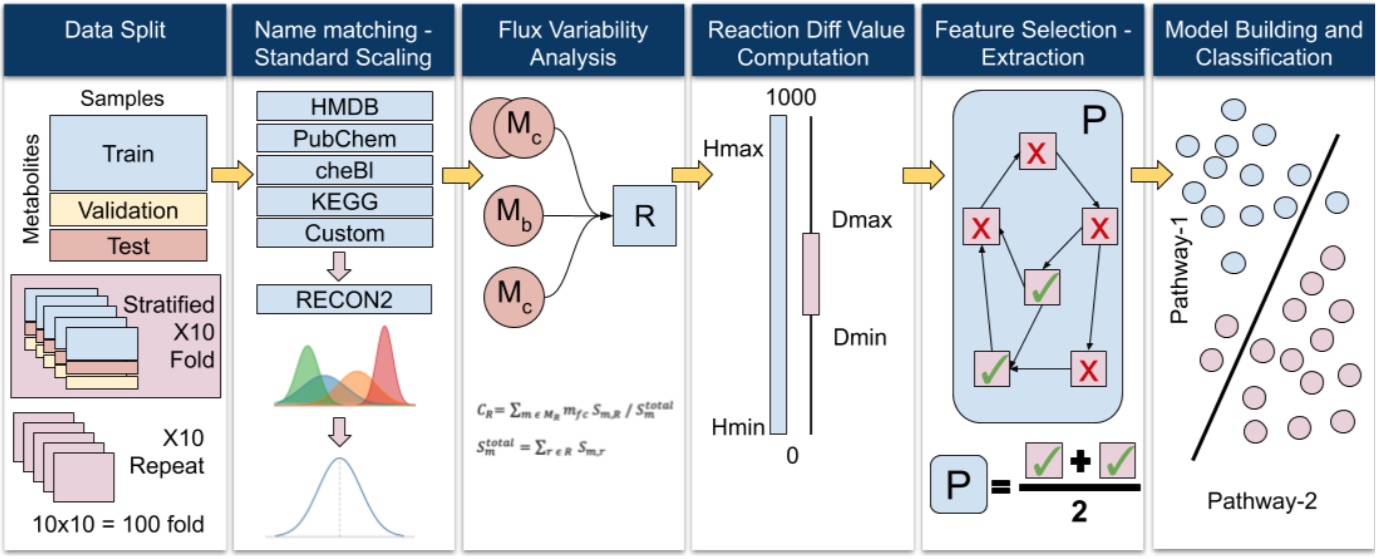
Metabolic networks consist of interacting biochemical reaction chains (i.e., pathways) that are responsible for essential cellular processes such as energy production, DNA synthesis, and lipid breakdown. Reactions in these networks are connected to each other through chemical substances (e.g., glucose) that they consume and produce. Such substances are called “metabolites”, and with the recent advancements in biotechnology, it is possible to simultaneously measure the concentrations for thousands of metabolites in biofluids (e.g., blood, urine, etc.). Extraordinary changes in metabolite concentrations often point to physiological conditions. Metabolomics is the study of these concentration changes as well as interpretation of what their biochemical implications may be. Interpreting the concentration changes of large numbers of metabolites with respect to a metabolic network is complicated, time consuming, and cumbersome. In order to decrease and manage the complexity, most researchers usually focus on only a small select set of metabolites among thousands. This usually results in omitting a large chunk of the big picture, and makes the analysis conclusions local and limited.
The goal of this project is to develop models, algorithms, and tools to automatically interpret large numbers of metabolite concentration changes in a holistic manner, and produce a set of biologically viable hypotheses explaining the observed changes. Besides, we aim to create health applications that will enable the use of the output from the above models and algorithms. More specifically, the goals of this project are as follows: (i) computationally model metabolomics data, and develop algorithms to make sense of the observed concentration changes in a biologically relevant way, (ii) associate the automatically created hypotheses with known physiological conditions, (iii) develop tools to enable medical doctors to use the results of such analyses in designing personalized treatment strategies for their patients, (iv) develop web-based analysis and visualization tools that will implement the above models and algorithms.
In order to achieve these goals, we propose the following approach: (a) Starting from observed metabolites as the only content of a metabolic subnetwork S, iteratively apply graph extension methods, and grow S with new reactions. At the end, each such subnetwork leads to a unique hypothesis showing active and inactive regions of the metabolic network that may have produced the observed changes. (b) At each extension point, we apply Flux Balance Analysis (Orth vd., 2010) locally on the reactions that involve the extension metabolite, and compute the fluxes of the newly added reactions. (c) Allosteric regulation and gene regulation relationships are checked to eliminate hypotheses that are biologically not viable. (d) The remaining hypotheses are checked whether they involve any overlapping parts with the known mechanisms for physiological conditions. If there are such overlaps, then the corresponding hypotheses are linked to those physiological conditions. In order to build the data infrastructure for all these analyses, we will establish our own database which will be populated from variety of data sources to include the metabolic network, gene regulation data, physiological conditions, their possible mechanisms, and the corresponding treatment strategies (if any). The developed methods and algorithms will be incorporated into web-based tools, and made publicly available to researchers and other interested users.
With the methods and tools proposed in this project, researchers will be able to take full advantage of having large numbers of metabolite measurements by quickly analyzing them as a whole with much less effort, rather than choosing a few metabolites and concentrating on their implications. In addition, the above discussed personalized medicine objective of the project will (i) enable medical doctors to make more informed treatment choices, (ii) prevent patients from going through trial of different alternative treatments so that they will not be exposed to unnecessary side effects of inappropriate treatments, and (iii) contribute to saving money by eliminating unnecessary treatments, and decreasing the amount of time that passes by until patients start receiving the right treatment. This would in turn minimize patients’ off time from work or personal business, and thus increase the total efficiency of the society.
Funding Institution: The Scientific and Technological Research Council of Turkey (TUBITAK CAREER 3501) (Role: PI, Duration: 21 Months, 2017 - 2019)
 New Methods and Algorithms to Estimate the Selectivity of SQL LIKE Queries
New Methods and Algorithms to Estimate the Selectivity of SQL LIKE Queries
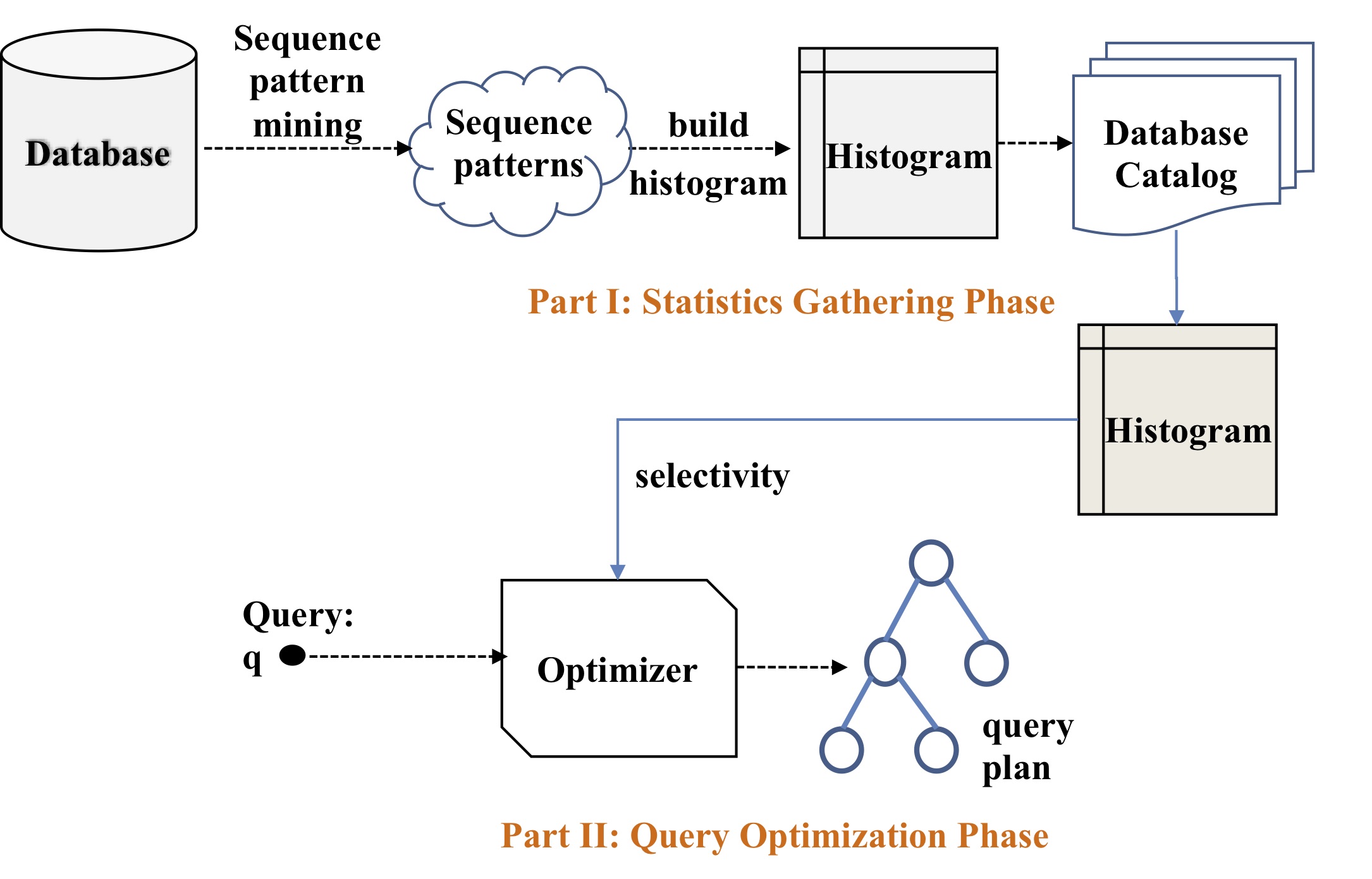
Accurate cost and time estimation of a query is one of the major success indicators for database management systems. SQL allows to express flexible queries on text-formatted data. The LIKE operator is used to search for a specified pattern in a string database (e.g., name LIKE ‘es%’ predicate allows to search for people whose names start with ‘es’). It is vital to estimate the selectivity of such flexible predicates accurately for the query optimizer to choose an efficient execution plan. In this project, we will study the problem of estimating the selectivity of a LIKE query predicate over a bag of strings.
We propose a new type of pattern-based histogram structure to summarize the data distribution in a particular column. More specifically, we will first mine sequential patterns over a given string database, and then construct a special histogram out of the mined patterns. During query optimization time, pattern-based histograms will be exploited to estimate the selectivity of a LIKE predicate. Besides, in this project, we will extend the existing sequence mining techniques to compute more specific sequence patterns and increase the selectivity estimation accuracy of the proposed framework. Orthogonal to the proposed techniques, as part of this project, we will question the value of the currently used metrics in the literature to compare different selectivity estimation methods. We argue that such methods are unnecessarily too specific, and may be misleading for the database practitioners and database management system developers. We will develop a new practical benefit-based metric, and provide an evaluation of the state of the art techniques as well as the methods that will be developed as part of this project. In this way, this project will provide a more realistic view of the alternative techniques to the researchers and developers in the field.
The proposed techniques will be assessed and compared to the state of the art in different dimensions on real and synthetic string datasets which are freely available to the researchers. The methods and techniques that will be developed during this project will enable database managements systems to come up with more optimal query execution plans by using less resources. Hence, flexible queries on text-based data will take less time to execute. Besides, during this project a graduate student will have the opportunity to get trained and prepare a thesis in this particular field. Through this project, we aim to submit two patent applications which will greatly contribute to the innovation capabilities of Turkey. Furthermore, the project results will be shared with the research community in the form of three different publications.
Funding Institution: The Scientific and Technological Research Council of Turkey (TUBITAK 1001) (Role: PI, Duration: 27 Months, 2014 - 2017)
 Data Mining Techniques for Efficient Query Optimization in Relational Database Management Systems
Data Mining Techniques for Efficient Query Optimization in Relational Database Management Systems
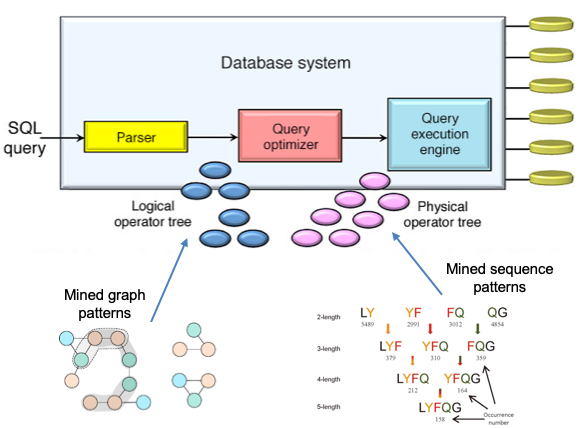
The query optimization module in a relational database management system is responsible for creating the most efficient query execution plan for a query. The number of alternative execution plans that need to be explored during query optimization time increases exponentially with the number of involved tables. However, the optimization is done at query compilation time. Hence, the optimization time is very limited. Therefore, early pruning of plan alternatives before they are evaluated is important to increase the performance of query optimization. The aim of this project is to improve the efficiency of query optimization using data mining techniques. More specifically, this research proposes that (i) exploiting graphical mining on query graphs for selecting appropriate query transformation during logical optimization, and (ii) eliminating high-cost table join order sequences based on the mined sequence patterns during physical optimization.
Funding Institution: The Scientific and Technological Research Council of Turkey (TUBITAK 2232) (Role: PI, Duration: 24 Months, 2014 - 2016)
|
|
|