PREPARATIVE
INFO (21.11.2002)
·
A unit mass very nearly equal to that of a hydrogen
atom (precisely equal to 1.0000 on the atomic mass scale). (p.24)
·
Named after John Dalton (1766-1844) who developed the
atomic theory of matter.
1 dalton = 1
atomic mass unit
1 kilodalton =
1000 dalton
1 dalton = 1.66x10-27
kg (www.xrefer.com)
1 dalton = 1 amu
ANGSTROM (Å)
·
A unit of length that is equal to 10-10 meter (p.28).
·
Named after Anders J. Ångström (1814-1874).
1 Å = 10-10
m = 10-8 cm = 10-4 mm = 10-1 nm
log (Å) = -0.0985
+ 0.4459*log(molecular weight in daltons)
R=0.9984
AP40 filter: 1.2
µm =12,000 Å = 2,340,000 kDa
0.45 µm filter:
4500 Å = 260,000 kDa
0.22 µm filter:
2200 Å = 52,000 kDa
|
Material (p.51) |
Mass unit |
|
|
Ribonuclease A |
12.4 kDa |
|
|
Myoglobin |
18 kDa |
|
|
Hemoglobin |
68 kDa |
|
|
Fibrinogen |
330 kDa |
|
|
LSU rRNA |
1,100 kDa |
|
|
E. coli RNA |
2,500 kDa |
|
|
Tobacco mosaic
virus |
40,000 kDa |
|
|
Bacterium |
1,550,000 kDa |
1000-5000 nm= (1-5)*104
Å |
|
Lysosome |
|
|
Ribonuclease A: Pancreatic
digestive enzyme, hydrolyzing RNA (p.35). A single polypeptide chain of 124
residues. Width is 20 Å.
Myoglobin: Protein carrying
oxygen in the muscle. Single compact polypeptide chain of 153 aminoacids.
Overall dimensions = 45*35*25 Å.
Amino acids: Mean molecular
weight of an a.a. residue isaround 110. Molecular weight of most polypeptide
chains is between 5,500 and 220,000.
Proteins: A protein having
a molecular weight of 50,000 has a mass of 50,000 dalton, or 50 kDa.
References:
1. Stryer, L.
(1995). Biochemistry. 4th Ed.
3. Brock, T.D.; et
al. (1994). Biology Microorgani
Membranes used by GUF_PSD
|
Filter Type |
Filter Size |
Max working presssure |
Storage |
Regeneration |
|
Filtration |
|
|
|
|
|
Millipore AP40, glass fiber |
1.2-1.6 m |
0.35 atm* |
Disposable |
Disposable |
|
DuraporeÒ HV, PVDF |
0.45 m |
0.35 atm* |
Disposable |
Disposable |
|
DuraporeÒ GV, PVDF |
0.22 m |
0.35 atm* |
Disposable |
Disposable |
|
Ultrafiltration |
|
|
|
|
|
Millipore, PL series |
100 kDa |
0.7 atm |
in 10 % EtOH, +4oC |
0.1 N NaOH, 30 min |
|
Millipore, PL series |
30 kDa |
3.7 atm |
in 10 % EtOH, +4oC |
0.1 N NaOH, 30 min |
|
Millipore, PL series |
10 kDa |
3.7 atm |
in 10 % EtOH, +4oC |
0.1 N NaOH, 30 min |
|
Millipore, PL series |
3 kDa |
3.7 atm |
in 10 % EtOH, +4oC |
0.1 N NaOH, 30 min |
|
Millipore, PL series |
1 kDa |
3.7 atm |
in 10 % EtOH, +4oC |
0.1 N NaOH, 30 min |
*No pressure recommendation is available from the
manufacturer for these disposable membranes. The value was estimated safely according
to the value available for 100 kDa membrane.
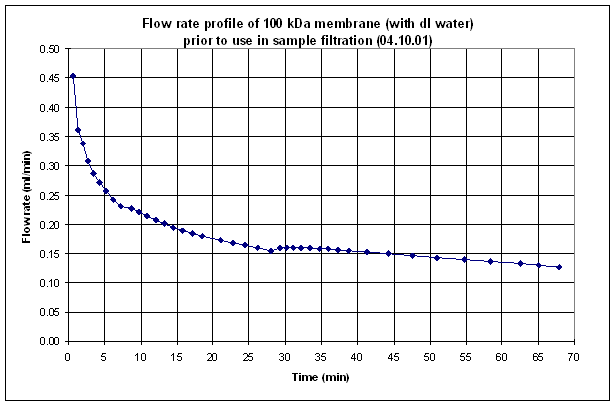
Preparative Work Prior to Starting the Experiments:
The filters,
especially the ultrafiltration membrane discs, should be checked in terms of
membrane rejection rate with filtration time, and thus against membrane
fouling. One way of doing this is thru running two experiments (one when using
the discs for the very first time prior to the experiments with actual samples,
and the other after filtering the samples from the discs) during which known
volumes of dI water are filtered and time of filtration is recorded. Thus, two
flow-rate profiles are obtained via plotting the recorded volumes of dI water
against time of filtration.
When the
difference between the flow-rate profiles before and after sample filtration is
significant (time point to reach the asymptotic flow-rate), the membrane discs
should be regarded exerting considerable membrane rejection, thus fouling, thus
the disc should be discarded and a new one should be used for further analyses.
All cleaning,
conditioning, and storing procedures should be performed in accordance with the
recommendations of the manufacturer, to help extend the time of efficient use
of the ultrafiltration membrane discs.
An example of a clean
flow-rate profile prior to use in sample filtration is given below for 100 kDa
membrane discs.